 |
|
Contact Information
Office: MEYR 475A
Phone: 410-455-1985
Email:
smitha@umbc.edu |
Associate Professor
Post-Doc, Northwestern University, 2016
M. Sc., Ph. D., University of Wisconsin—Madison, 2012
B. A., Boston University, 2007
Professional Interests
Research in my lab focuses on understanding the structure and function of metalloproteins involved in crucial biological processes such as protein translation, peptide posttranslational modification, nutrient uptake, and protein regulation. My laboratory sits at the interface of chemistry, biochemistry, and biophysics, and members of my lab have the opportunity to learn a wide array of techniques from molecular biology and protein expression and purification (both soluble and membrane), to structural biology, spectroscopy, and anaerobic techniques. We welcome chemists, biochemists, and molecular biologists alike who are interested in understanding the role of inorganic elements in biological settings!
There are two current projects in the lab: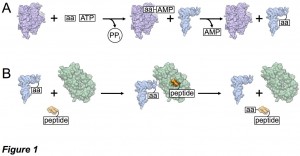
Protein Translation, Posttranslational Modification, and Heme
Translation is the process by which genetic information in the form of messenger ribonucleic acid (mRNA) is decoded into a synthesized string of amino acids corresponding to a protein. Transfer RNAs (tRNAs) are single- stranded RNA molecules chiefly responsible for carrying and delivering amino acids to the ribosome for polypeptide synthesis, and tRNAs are aminoacylated in an energy-dependent manner at their 3’ end by a corresponding aminoacyl-tRNA synthetase (aaRS) (Figure 1A).
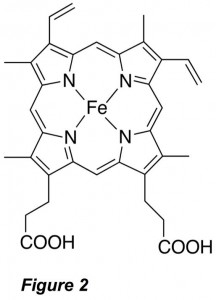 Besides their role in protein translation, aminoacylated tRNAs may be utilized for novel antibiotic synthesis, pathogenic bacterial membrane modification and cell wall synthesis, posttranslational peptide modifications and protein degradation, and even cofactor biosynthesis. For these uses, most organisms employ an aminoacyl tRNA transferase (aa-transferase) that functions to transfer the amino acid from the tRNA to a protein or peptide (Figure 1B). Intriguingly, both the aminoacyl-tRNA synthetase and aminoacyl tRNA transferase that function with the amino acid arginine (Arg) as their substrates appear to be regulated by iron protoporphyrin IX (heme b; Figure 2). These observations suggest a connection between protein biosynthesis, protein degradation, and in vivo supplies of Arg, the precursor to the gaseous neutrotransmitter and vasodilator nitric oxide (NO). However, the structural and mechanistic underpinnings of these processes remain poorly understood. We are using structural, spectroscopic, and enzymatic approaches to understand the mechanism of these enzymes and how regulatory elements control their function.
Besides their role in protein translation, aminoacylated tRNAs may be utilized for novel antibiotic synthesis, pathogenic bacterial membrane modification and cell wall synthesis, posttranslational peptide modifications and protein degradation, and even cofactor biosynthesis. For these uses, most organisms employ an aminoacyl tRNA transferase (aa-transferase) that functions to transfer the amino acid from the tRNA to a protein or peptide (Figure 1B). Intriguingly, both the aminoacyl-tRNA synthetase and aminoacyl tRNA transferase that function with the amino acid arginine (Arg) as their substrates appear to be regulated by iron protoporphyrin IX (heme b; Figure 2). These observations suggest a connection between protein biosynthesis, protein degradation, and in vivo supplies of Arg, the precursor to the gaseous neutrotransmitter and vasodilator nitric oxide (NO). However, the structural and mechanistic underpinnings of these processes remain poorly understood. We are using structural, spectroscopic, and enzymatic approaches to understand the mechanism of these enzymes and how regulatory elements control their function.
 Pathogenic Iron Uptake and Delivery:
Pathogenic Iron Uptake and Delivery:
Iron is an essential element for virtually every living organism and has been adopted to serve in major biological processes such as nitrogen fixation, methane oxidation, hydrogen production, aerobic cellular respiration, oxygen transport, DNA biosynthesis, and even gene regulation. However, iron-based life represents a double-edged sword, as Fe(II) is bioavailable but highly reactive, whereas Fe(III) is intractable but fairly chemically inert. Every organism that utilizes iron employs biological pathways to obtain this element from the environment, to regulate its bioavailable concentration, and to sequester its excess. In order to establish infection in humans, pathogenic bacteria have evolved several responses to manage iron acquisition and utilization. In particular, sophisticated high-affinity metal-transport systems enable pathogens to scavenge iron from their surroundings. The ability to acquire and utilize iron aids in the proliferation of several pernicious bacteria, such as Heliobacter pylori (ulcer-causing agent), Legionella pneumophila (causative agent of Legionnaire’s disease), Campylobacter jejuni (common cause of food poisoning), Klebsiella pneumoniae(common cause of nosocomial infections), Vibrio cholerae (causative agent of cholera), and Salmonella enterica (another common cause of food poisoning).
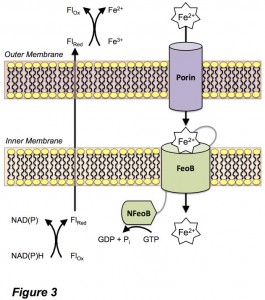
One important route for the uptake of Fe(II) for bacteria is via the ferrous iron uptake (Feo) system (Figure 3). However, the mechanism of iron uptake through this pathway is poorly understood by comparison to Fe(III) and heme uptake pathways. In an era of increasing antibacterial resistance, understanding and targeting the routes of bacterial nutrient uptake are crucial to stem bacterial virulence. We are using a multifaceted approach to understand the structure and function of proteins involved in bacterial Fe(II) uptake.
Selected Publications
Cartwright, M.; Van, V.; and Smith, A. T. The preparation of recombinant arginyltransferase 1 (ATE1) for biophysical characterization. Methods Enzymol. 2022, 678, In press. DOI: 10.1016/bs.mie.2022.07.036
Sestok, A. E.; O’Sullivan, S. M.#; and Smith A. T. A general protocol for the expression and purification of the intact transmembrane transporter FeoB. BBA-Biomembranes, 2022, 1864, 183973.
Sestok, A. E.; Brown, J. B.; Obi, J. O.; O’Sullivan, S. M.; Garcin, E. D.; Deredge, D. J.; and Smith A. T. A fusion of the Bacteroides fragilis ferrous iron import proteins reveals a role for FeoA in stabilizing GTP-bound FeoB. J. Biol. Chem. 2022, 298, 101808.
Brown, J. B.; Lee, M. A.; and Smith, A. T. The structure of Vibrio chloerae FeoC reveals conservation of the helix-turn-helix motif but not the cluster-binding domain. J. Biol. Inorg. Chem. 2022, 27, 485-495 and bioRxiv. 2022, DOI: 10.1101/2022.02.26.482101
Sestok, A. E.; Lee, M.; and Smith, A. T. Prokaryotic ferrous iron uptake: exploiting pools of reduced iron across multiple microbial environments. In: Hurst, C.J. (eds) Microbial Metabolism of Metals and Metalloids. Advances in Environmental Microbiology, 2022, 10, 299-357. Springer, Cham. https://doi.org/10.1007/978-3-030-97185-4_12
Smith, A. T. Protein arginylation is regulated during SARS-CoV-2 infection. Rapid Reviews: COVID-19. 2022, DOI: 10.1162/2e3983f5.63d125d7
Sestok, A. E.; Brown, J. B.; Obi, J. O.; O’Sullivan, S. M.; Garcin, E. D.; Deredge, D. J.; and Smith A. T. Biochemical and structural characterization of the fused Bacteroides fragilis NFeoAB domain reveals a role for FeoA. bioRxiv. 2021. DOI: https://doi.org/10.1101/2021.09.29.462438
Sánchez-Osuna, M.; Cortés, P.; Lee, M.; Smith, A. T.; Barbé, J.; and Erill, I. Non-canonical LexA proteins regulate the SOS response in the Bacteroidetes. Nucleic Acids Res. 2021, 49, 11050-11066.
Van, V.; Brown, J. B.; Rosenbach, H.; Mohamed, I.#; Ejimogu, N.-E.#; Bui, T. S.#; Szalai, V. A.; Chacón, K. N.; Span, I.; and Smith, A. T. Iron-sulfur clusters are involved in post-translational arginylation. bioRxiV. 2021. DOI: https://doi.org/10.1101/2021.04.13.439645
Cain, T. J. and Smith, A. T. Ferric iron reductases and their contribution to unicellular ferrous iron uptake. J. Inorg. Biochem. 2021, 218, 11407, 1-9.
Smith, A. T.; Linkous, R. O.#; Max, N. J.; Sestok, A. E.; Szalai, V.A.; and Chacón, K. N. Escherichia coli FeoC binds a redox-active, rapidly oxygen-sensitive [4Fe-4S] cluster. ChemRxiv. 2019. DOI: https://doi.org/10.26434/chemrxiv.8411408.v1
Linkous, R. O.#; Sestok, A. E.; and Smith, A. T. The crystal structure of Klebsiella pneumoniae FeoA reveals a site for protein-protein interactions. Proteins. 2019, 87, 897-903. DOI:https://doi.org/10.1002/prot.25755 and bioRxiv. 2019. DOI: https://doi.org/10.1101/514059
Sestok, A. E.; Linkous, R. O.#; and Smith, A. T. Toward a Mechanistic Understanding of Feo-Mediated Ferrous Iron Uptake. Metallomics. 2018, 10, 887-898.†
Smith, A. T. and Sestok, A. E. Expression and purification of functionally active ferrous iron transporter FeoB from Klebsiella pneumoniae. Protein Expr. Purif. 2018, 142, 1-7.
Smith, A. T.; Ross, M. O.; Hoffman, B. M.; Rosenzweig, A. C. Metal Selectivity of a Cd-, Co-, and Zn-Transporting P1B-type ATPase. Biochemistry 2017, 56, 85-95.
Hines, J. P.; Smith, A. T.; Jacob, J. P.; Lukat-Rodgers, G. S.; Barr, I.; Rodgers, K. R.; Guo, F.; Burstyn, J. N. CO and NO bind to Fe(II) DiGeorge critical region 8 heme but do not restore primary microRNA processing activity. J. Biol. Inorg. Chem. 2016, 21, 1021-1035.
Kathman, S.; Span, I.; Smith, A. T.; Xu, Z.; Zhan, J.; Rosenzweig, A. C.; Statsyuk, A. Discovery and structural characterization of covalent inhibitors of Nedd4-1 ubiquitin ligase processivity J. Am. Chem. Soc. 2015, 137, 12442-12445.
Smith, A. T. * ; Pazicni, S. * ; Marvin, K. A. * ; Stevens, D. J.; Freeman, K. M.; Burstyn, J. N. Functional divergence of heme-thiolate proteins: a classification based on spectroscopic attributes. Chem. Rev. 2015, 115, 2532-2558.
Smith, A. T., Smith, K.P., and Rosenzweig, A. C. Diversity of the metal-transporting P1B-type ATPases. J. Biol. Inorg. Chem. 2014, 19, 947-960.
Smith, A. T.; Marvin, K. A.; Freeman, K. M.; Kerby, R. L.; Roberts, G. P.; Burstyn, J. N. Identification of Cys94 as the distal ligand to the Fe(III) heme in the transcriptional regulator RcoM-2 from Burkholderia xenovorans. J. Biol. Inorg. Chem. 2012, 17, 1071-1082.
Smith, A. T.; Su, Y.; Stevens, D. J.; Majtan, T.; Kraus, J. P.; Burstyn, J. N. The effect of the disease-causing R266K mutation on the heme and PLP environments of the human enzyme cystathionine β-synthase. Biochemistry 2012, 51, 6360-6370.
Barr, I.; Smith, A. T.; Chen, Y.; Senturia, R.; Burstyn, J. N.; Guo, F. Ferric, not ferrous, heme activates RNA-binding protein DGCR8 for primary microRNA processing. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 1919-1924.
Barr, I.; Smith, A. T.; Senturia, R.; Chen, Y.; Burstyn, J. N.; Guo, F. DiGeorge Critical Region 8 (DGCR8) is a double-cysteine ligated heme protein. J. Biol. Chem.2011, 286, 16716-16725.
Smith, A. T.; Majtan, T.; Freeman, K. M.; Su, Y.; Kraus, J. P.; Burstyn, J. N. Cobalt cystathionine β-synthase: a cobalt-substituted heme protein with a unique thiolate ligation motif. Inorg. Chem. 2011, 50, 4417-4427.
Matjan, T.; Freeman, K. M.; Smith, A. T.; Burstyn, J. N.; Kraus, J. P. Purification and characterization of cystathionine β-synthase bearing cobalt protoporphyrin. Arch. Biochem. Biophys. 2011, 508, 25-30.
†Highlighted as the journal cover.
#Indicates undergraduate author.
Honors and Awards
NSF CAREER Award (UMBC) 2019
American Heart Association Career Development Grant (UMBC) 2019
Career Center Impact Recognition Award (UMBC) 2018
Summer Faculty Fellowship (UMBC) 2017
NIH Ruth L. Kirschstein NRSA Postdoctoral Fellowship (NU) 2013
University Housing’s Honored Instructor Award (UW) 2011
Charles & Martha Casey Excellence in Research Award (UW) 2011
Outstanding Chemistry Teaching Award (UW) 2010
Undergraduate Mentoring Award (UW) 2010
Undergraduate Research Award (BU) 2007
Department of Chemistry Research Award (BU) 2007
CHEMIA Vice President (BU) 2005—2007
Mark Riemen Summer Research Prize (BU) 2006
Courses Taught
CHEM437 Comprehensive Biochemistry I
Spring 2020
CHEM406/606 Bioinorganic Chemistry
GPLS709 Advanced Biochemistry
Fall 2019
CHEM437 Comprehensive Biochemistry I
Spring 2019
GPLS709 Advanced Biochemistry
Fall 2018
CHEM437 Comprehensive Biochemistry I
Spring 2018
CHEM490/606 Bioinorganic Chemistry
GPLS709 Advanced Biochemistry
Fall 2017
CHEM437 Comprehensive Biochemistry I
Spring 2017
GPLS709 Advanced Biochemistry
Fall 2016
CHEM490/606 Bioinorganic Chemistry
