 |
|
CONTACT INFORMATION
Office: CHEM 475A
Phone: 410-455-2510
Email: karpel@umbc.edu
|
Professor
Professional Interests
Structure-function studies on single-stranded nucleic acid binding proteinsI am predominantly interested in structure-function studies on single-strand specific nucleic acid binding proteins (also known as helix-destabilizing proteins). As a class, the binding proteins have a wide range of biological roles, yet they share a number of biochemical commonalities. These include homologous functional domains, regions responsible for nucleic acid interaction, binding cooperativity, and interaction with other proteins.
The binding proteins under current investigation include bacteriophage T4 gene 32 protein, involved in DNA replication, recombination and repair, and the nucleocapsid (NC) proteins of retroviruses, which have a role in viral RNA packaging and replication, and act as “chaperones”, facilitators of nucleic acid conformational change. Gene 32 protein, historically one of the first of this class of proteins to be characterized, continues to provide us with fascinating insights.
 |
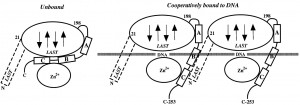
| Structure of gene 32 protein core domain bound to a single-stranded oligonucleotide |
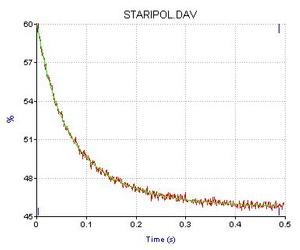 |
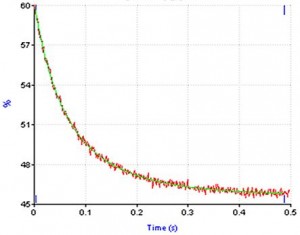
| Typical stopped-flow trace of gene 32 protein *I truncate binding poly(dT) |
Having identified an amino acid sequence (the‘LAST’ Motif) involved in both protein-nucleic acid and protein-protein interaction, we are particularly concerned with understanding the relationship of these two binding activities, and in further delineating the structural basis of the protein’s binding cooperativity and nucleic acid binding surface, the kinetic control of helix-destabilizing activity, and the protein’s interaction with other T4 replication, repair, and recombination proteins. In our structure-function studies we employ a broad spectrum of experimental approaches, including biophysical, protein chemical, and molecular biological methodologies.
We recently initiated nucleic acid binding studies on crotamine, a toxic, basic protein that is a major component of the venom of the South American rattlesnake. Crotamine has been shown to be a cell-penetrating protein that localizes in the chromosome. It has potential use as a drug-delivery vehicle. At present nothing is known about the DNA- and RNA-interactive properties of crotamine, and we present several approaches toward quantifying these interactions.
Selected Publications
- R.L. Karpel, DNA Binding Proteins and Drug Delivery Vehicles: Tales of Elephants and Snakes, Current Protein & Peptide Science, 16, 718-726 (2015).
- C. Macazo, R. L. Karpel, R. J. White, Monitoring Cooperative Binding using Electrochemical DNA-Based Sensors, Langmuir, 31, 868-875 (2015).
- L. Karpel, The Illusive Search for the Lowest Free Energy State of a Globular Protein, DNA Repair 21, 158-162 (2014).
- E. S. Yamane, F. C. Bizerra, E. B. Oliveira, M. Rajabi , G. L.C. Nunes, A. O. de Souza, K. Konno, I. D.C.G. da Silva, T. Yamane, R. L. Karpel, P. I. Silva Jr., and M. A. F. Hayashi, Unraveling the antifungal activity of a South American rattlesnake toxin crotamine, Biochimie, 95, 231-240 (2013).
- P.-C. Chen, M. A. F. Hayashi, E. Brandt Oliveira, and R. L. Karpel, DNA-Interactive Properties of Crotamine, a Cell-Penetrating Polypeptide and a Potential Drug Carrier, PLOS One, 7 (11), e48913 (2012).
- M. A. F. Hayashi, E. B. Oliveira, Irina Kerkis, R. L. Karpel, Crotamine, a Novel Cell-Penetrating Polypeptide Nanocarrier with Potential Anti-Cancer and Biotechnological Applications, Methods Mol Biol., 906, 337-52, (2012).
- M.C. Williams, I. Rouzina, and R.L. Karpel, Quantifying DNA–Protein Interactions by Single Molecule Stretching, Methods in Cell Biology, 84, 517-540 (2008).
- M.C. Williams, I. Rouzina, and R.L. Karpel, Thermodynamics and kinetics of DNA-protein interactions from single molecule force spectroscopy measurements, Current Organic Chemistry, 10, 419-432 (2006).
- I. Rouzina, K. Pant, R.L. Karpel, and M. C. Williams, Theory of electrostatically-regulated binding of T4 gene 32 protein to single- and double-stranded DNA, Biophys. J., 89, 1941-1956 (2005).
- K. Pant, R.L. Karpel, I. Rouzina, and M. C. Williams, Salt dependent binding of T4 gene 32 protein to single- and double-stranded DNA: Single molecule force spectroscopy measurements, J. Mol. Biol.,349, 317-330 (2005).
- M C. Williams, K. Pant, I. Rouzina, and R. L. Karpel. Single molecule force spectroscopy studies of DNA denaturation by T4 gene 32 protein, Spectroscopy, An International Journal, 18, 203-210 (2004).
- K. Pant, R.L. Karpel, I. Rouzina, and M.C. Williams, Mechanical measurement of single molecule binding rates: Kinetics of DNA helix-destabilization by T4 gene 32 protein, J. Mol. Biol., 336, 851-870 (2004).
- K. Pant, R.L. Karpel, and M.C. Williams, Kinetic Regulation of single DNA molecule denaturation by T4 gene 32 protein structural domains, J. Mol. Biol., 327, 571-578 (2003).
- R.L. Karpel, LAST Motifs and SMART Domains in Gene 32 Protein: An Unfolding Story of Autoregulation?,IUBMB Life, 53, 161-166 (2002).
- M.A. Urbaneja, M. Wu, J.R. Casas-Finet and R.L. Karpel, HIV-1 Nucleocapsid Protein as a Nucleic Acid Chaperone: Spectroscopic Study of its Helix-Destabilizing Properties, Structural Binding Specificity, and Annealing Activity, J. Mol. Biol., 318, 749-764 (2002).
- R.L. Karpel, Prokaryotic DNA Binding Proteins, Article 1042 in Encyclopedia for Life Sciences, vol. 15, pp. 231-238, Macmillan, 2002.
- L.A. Waidner, E.K. Flynn, M. Wu, X. Li, and R.L. Karpel, Domain Effects on the DNA-Interactive Properties of Bacteriophage T4 Gene 32 Protein, J. Biol. Chem., 276, 2509-2516 (2001).
- M. Wu, E.K. Flynn and R.L. Karpel , Details of the Nucleic Acid Binding Site of T4 Gene 32 Protein Revealed by Proteolysis and DNA Tm Depression Methods, J.Mol.Biol. 286, 1107-1121 (1999).
- J. Zaia, D. Fabris, D. Wei, R.L. Karpel and C. Fenselau, Monitoring metal ion flux in reactions of metallothionein and drug-modified metallothionein by electrospray mass spectrometry, Protein Science, 7, 2398-2404 (1998).
- D. Herschlag, M. Khosla, Z. Tsuchihashi and R.L. Karpel, An RNA chaperone activity of nonspecific RNA binding proteins in hammerhead ribozyme catalysis, EMBO J., 13, 2913-2924 (1994).
- M.D. Delahunty, S.H. Wilson and R.L. Karpel, Studies on primer binding of HIV-1 reverse transcriptase using a fluorescent probe, J. Mol. Biol. 236, 469-479 (1994).
- J.R. Casas-Finet and R.L. Karpel, Bacteriophage T4 gene 32 protein: Modulation of protein-nucleic acid and protein_protein association by structural domains, Biochemistry, 32, 9735-9744 (1993).
- J.R. Casas-Finet, J.D. Smith, Jr., A. Kumar, J.G. Kim, S.H. Wilson and R.L. Karpel, Mammalian hnRNP A1 and its Constant Domains: Self-Association, Structural Stability, and Binding Cooperativity and Specificity, J. Mol. Biol., 229, 873-889 (1993).
- J.R. Casas-Finet, A. Kumar, R.L. Karpel and S.H. Wilson, Mammalian DNA Polymerase-β. Characterization of a 16-K Da Transdomain Fragment Containing the Nucleic Acid Binding Activities of the Native Enzyme,Biochemistry 31, 10272-10280 (1992).
- J.R. Casas-Finet, S.H. Wilson and R.L. Karpel, Selective Photochemical Modification by Trichloroethanol of Tryptophan Residues in Proteins with a High Tyrosine-to-Tryptophan Ratio, Analytical Biochemistry, 205, 27-35 (1992).
- M.D. Delahunty, T.L. South, M.F. Summers and R.L. Karpel, Nucleic Acid Interactive Properties of a Peptide Corresponding to the N-terminal Zinc Finger Domain of HIV-1 Nucleocapsid Protein, Biochemistry, 31, 6461-6469 (1992).
- J.R. Casas-Finet, K.R. Fischer, and R.L. Karpel, Structural Basis for the Nucleic Acid Binding Cooperativity of Bacteriophage T4 Gene 32 Protein: The (Lys/Arg)3(Ser/Thr)2 (LAST) Motif, Proc. Natl. Acad. Sci. USA, 89, 1050-1054 (1992).
- J.R. Casas-Finet, R.L. Karpel, A.H. Maki, A. Kumar and S.H. Wilson, Physical Studies of Tyrosine and Tryptophan Residues in Mammalian A1 Heterogeneous Nuclear Ribonucleoprotein: Evidence for a Segmented Structure, J. Mol. Biol., 221, 693-709 (1991).
- A. Kumar, J.R. Casas-Finet, C.J. Luneau, R.L. Karpel, B. Merrill, K.R. Williams and S.H. Wilson, Mammalian Heterogeneous Nuclear Ribonucleoprotein A1: Nucleic Acid Binding Properties of the COOH-Terminal Domain, J. Biol. Chem. 265, 17094-17100 (1990).
Research Description
 |
 |
Structure-function studies on single-stranded nucleic acid binding proteins
I am interested in structure-function studies on single-strand specific nucleic acid binding proteins (also known as helix-destabilizing proteins). As a class, the binding proteins have a wide range of biological roles, yet they share a number of biochemical commonalities. These include homologous functional domains, regions responsible for nucleic acid interaction, binding cooperativity, and interaction with other proteins.
The binding proteins under current investigation include bacteriophage T4 gene 32 protein, involved in DNA replication, recombination and repair, and the nucleocapsid (NC) proteins of retroviruses, which have a role in viral RNA packaging and replication, and act as “chaperones”, facilitators of nucleic acid conformational change. Gene 32 protein, historically one of the first of this class of proteins to be characterized, continues to provide us with fascinating insights.
We are particularly concerned with the structural basis of binding cooperativity, the control of helix-destabilizing activity, and the interaction with other T4 replication, repair, and recombination proteins.
In our structure-function studies we employ a broad spectrum of experimental approaches, including biophysical, protein chemical, and molecular biological methodologies.